【DoubletFinder】predicts doublets in single-cell RNA sequencing data
remotes::install_github('chris-mcginnis-ucsf/DoubletFinder')DF共分成四步:
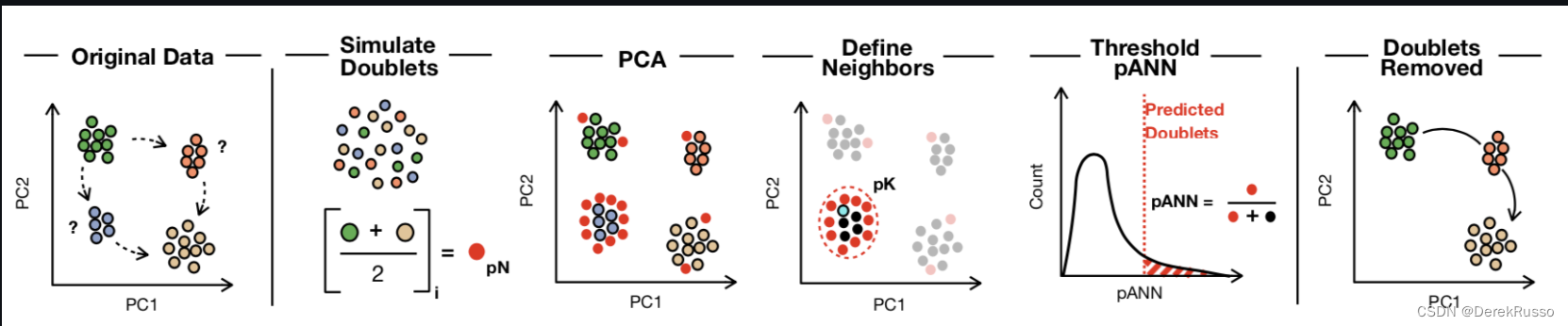

需要输入的参数:
(1)seu ~ fully processed seurat object (i.e., after NormalizeData, FindVariableGenes, ScaleData, RunPCA, and RunTSNE have all been run)
(2)PCs ~ The number of statistically-significant principal components, specified as a range (e.g., PCs = 1:10)
(3)pN ~ This defines the number of generated artificial doublets, expressed as a proportion of the merged real-artificial data. Default is set to 25%
(4)pK ~ This defines the PC neighborhood size used to compute pANN, expressed as a proportion of the merge
本文来自互联网用户投稿,文章观点仅代表作者本人,不代表本站立场,不承担相关法律责任。如若转载,请注明出处。 如若内容造成侵权/违法违规/事实不符,请点击【内容举报】进行投诉反馈!