单细胞三大R包之scater
1.准备R包和数据
library(BiocStyle)
set.seed(10918)
library(scRNAseq)
example_sce <- ZeiselBrainData()
example_sce## class: SingleCellExperiment
## dim: 20006 3005
## metadata(0):
## assays(1): counts
## rownames(20006): Tspan12 Tshz1 ... mt-Rnr1 mt-Nd4l
## rowData names(1): featureType
## colnames(3005): 1772071015_C02 1772071017_G12 ... 1772066098_A12
## 1772058148_F03
## colData names(10): tissue group # ... level1class level2class
## reducedDimNames(0):
## altExpNames(2): ERCC repeat# 可以看到维度20006 3005
2.质控
质控,去掉线粒体基因(以mt开头的)
library(scater)
example_sce <- addPerCellQC(example_sce, subsets=list(Mito=grep("mt-", rownames(example_sce))))
3.了解细胞现有的注释信息
coldata里面有记录tissue和class
tmp = as.data.frame(example_sce@colData)
table(tmp$tissue)##
## ca1hippocampus sscortex
## 1314 1691table(tmp$level1class)##
## astrocytes_ependymal endothelial-mural interneurons
## 224 235 290
## microglia oligodendrocytes pyramidal CA1
## 98 820 939
## pyramidal SS
## 399table(tmp$sex)##
## 1 2 3
## 1524 46 1435fivenum(tmp$total.mRNA.mol)## [1] 1 703 1429 2144 2831#横坐标是总count数,纵坐标是检测到的基因数量。
plotColData(example_sce, x = "sum", y="detected", colour_by="tissue")
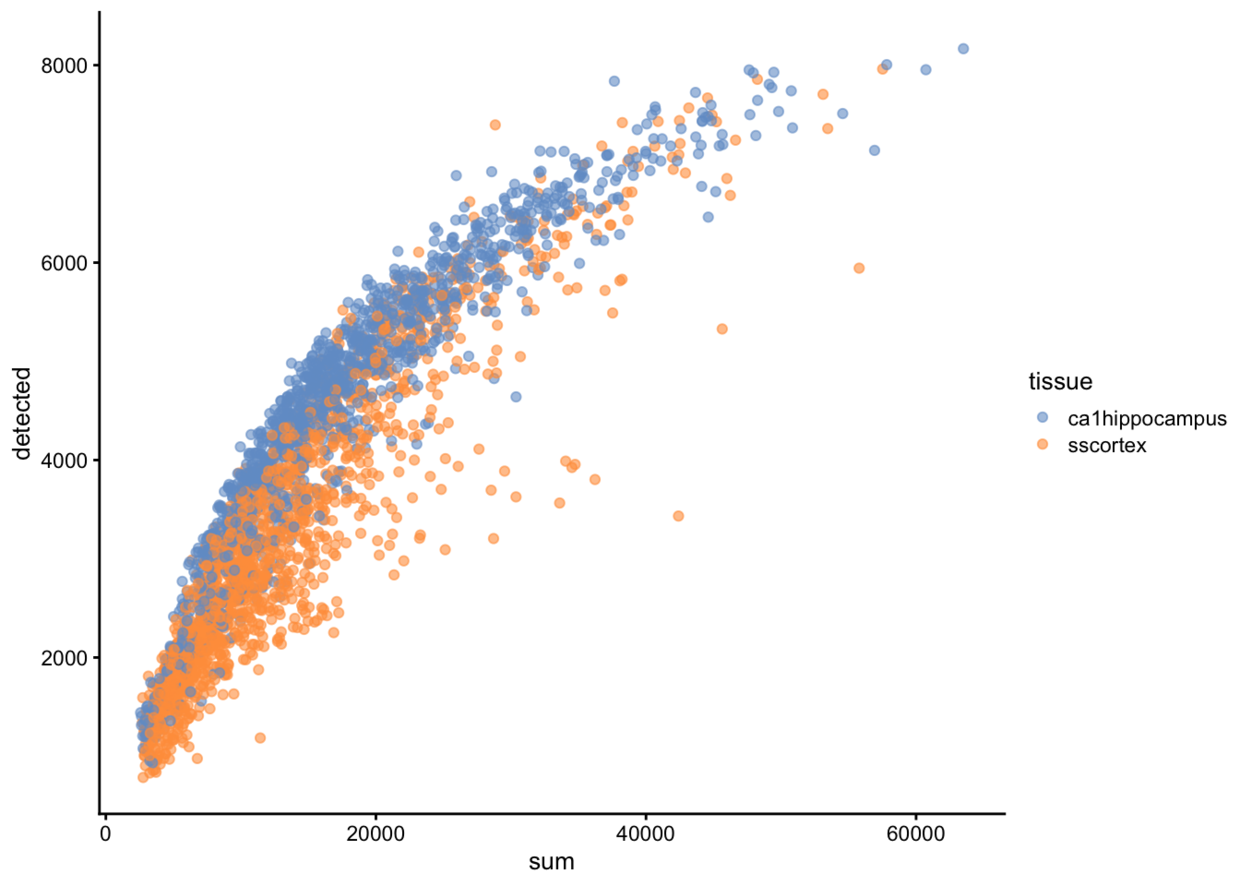
#线粒体基因的比例
plotColData(example_sce, x = "sum", y="subsets_Mito_percent",other_fields="tissue") + facet_wrap(~tissue)
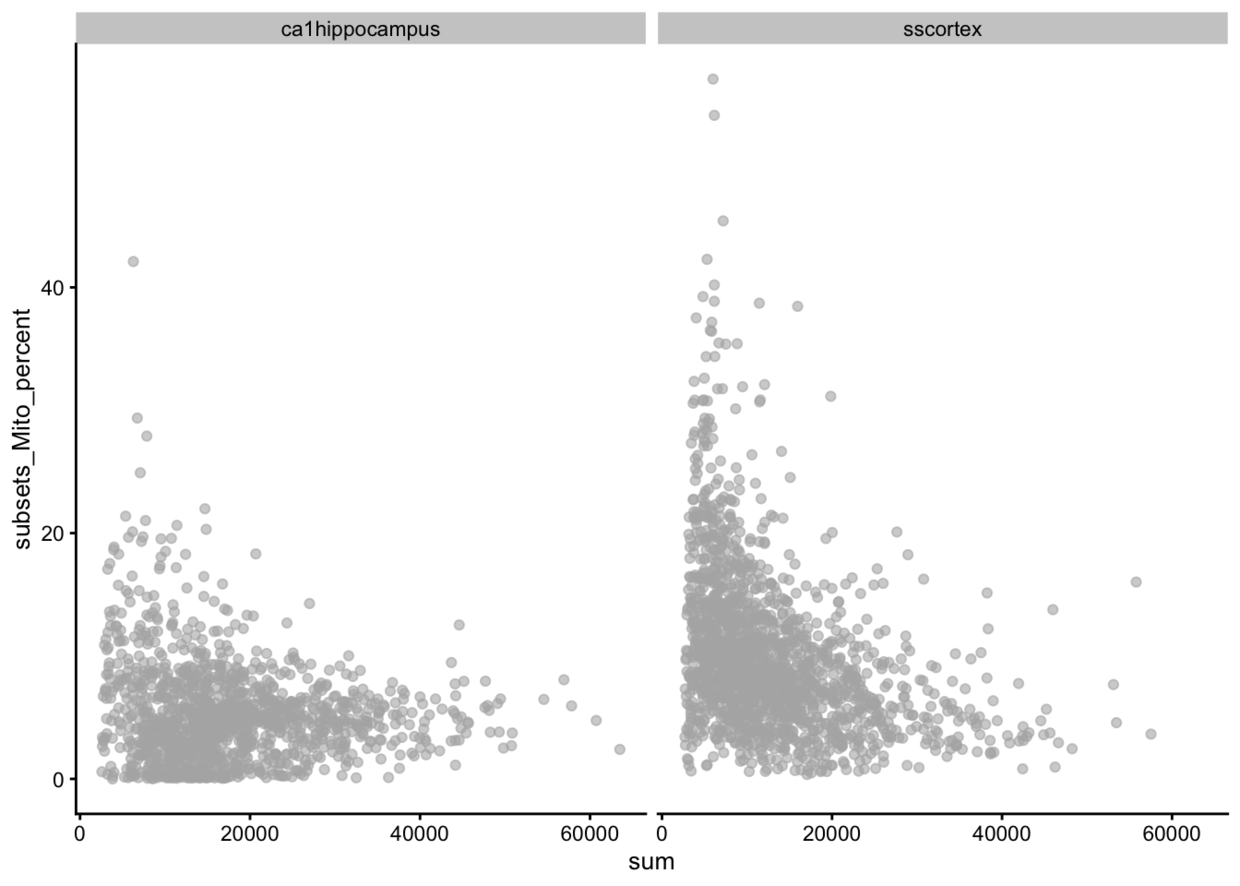
按照tissue分了两张子图。
看每个变量对解释变异的贡献
example_sce <- logNormCounts(example_sce) vars <- getVarianceExplained(example_sce, variables=c("tissue", "total mRNA mol", "sex", "age"))
head(vars)## tissue total mRNA mol sex age
## Tspan12 0.02207262 0.074086504 0.146344996 0.09472155
## Tshz1 3.36083014 0.003846487 0.001079356 0.31262288
## Fnbp1l 0.43597185 0.421086301 0.003071630 0.64964174
## Adamts15 0.54233888 0.005348505 0.030821621 0.01393787
## Cldn12 0.03506751 0.309128294 0.008341408 0.02363737
## Rxfp1 0.18559637 0.016290703 0.055646799 0.02128006plotExplanatoryVariables(vars)
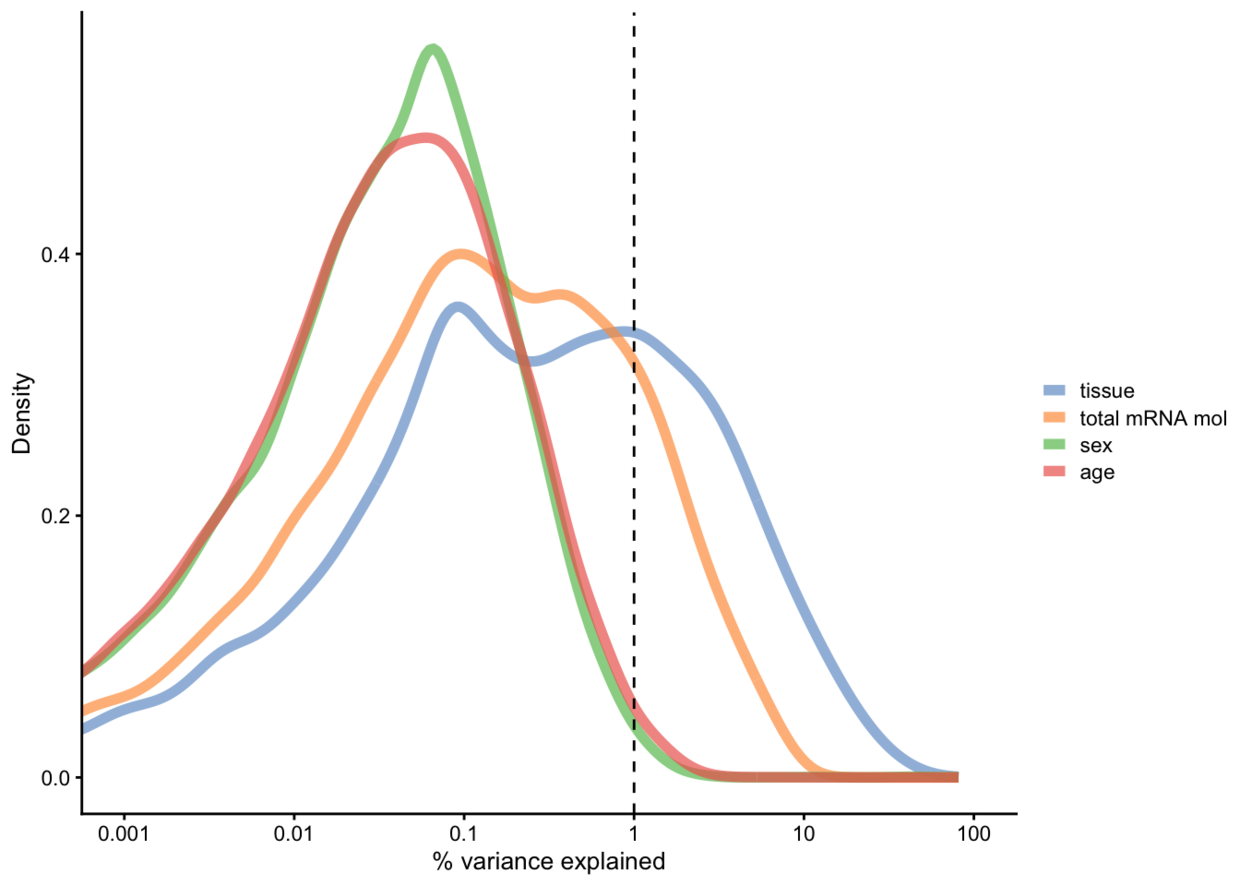
画基因表达量与注释信息关系
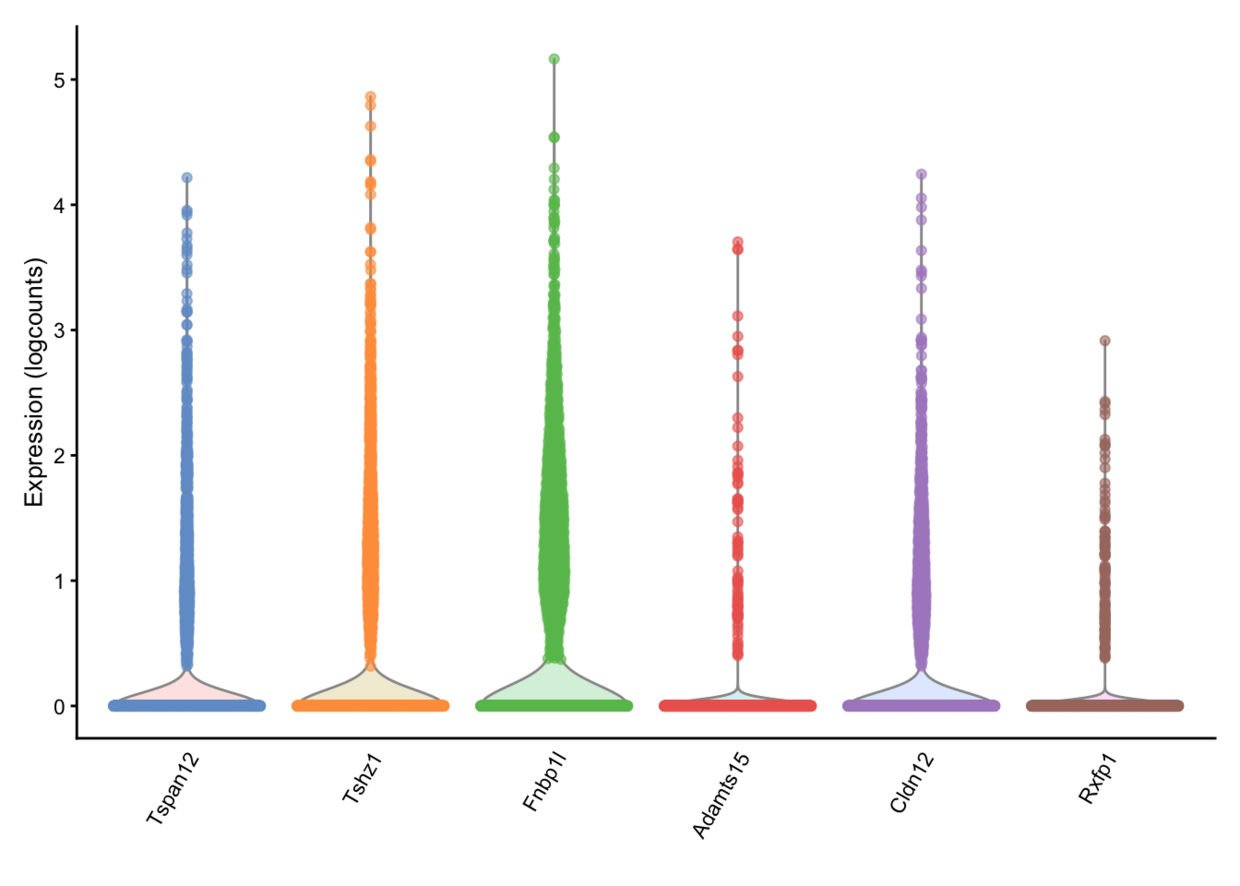
x是协变量,如果是个连续型变量,就画出点图,是离散型变量,则画小提琴图。如果不指定x,就每个基因成为一个小提琴图。
plotExpression(example_sce, rownames(example_sce)[1:6],x = rownames(example_sce)[10])
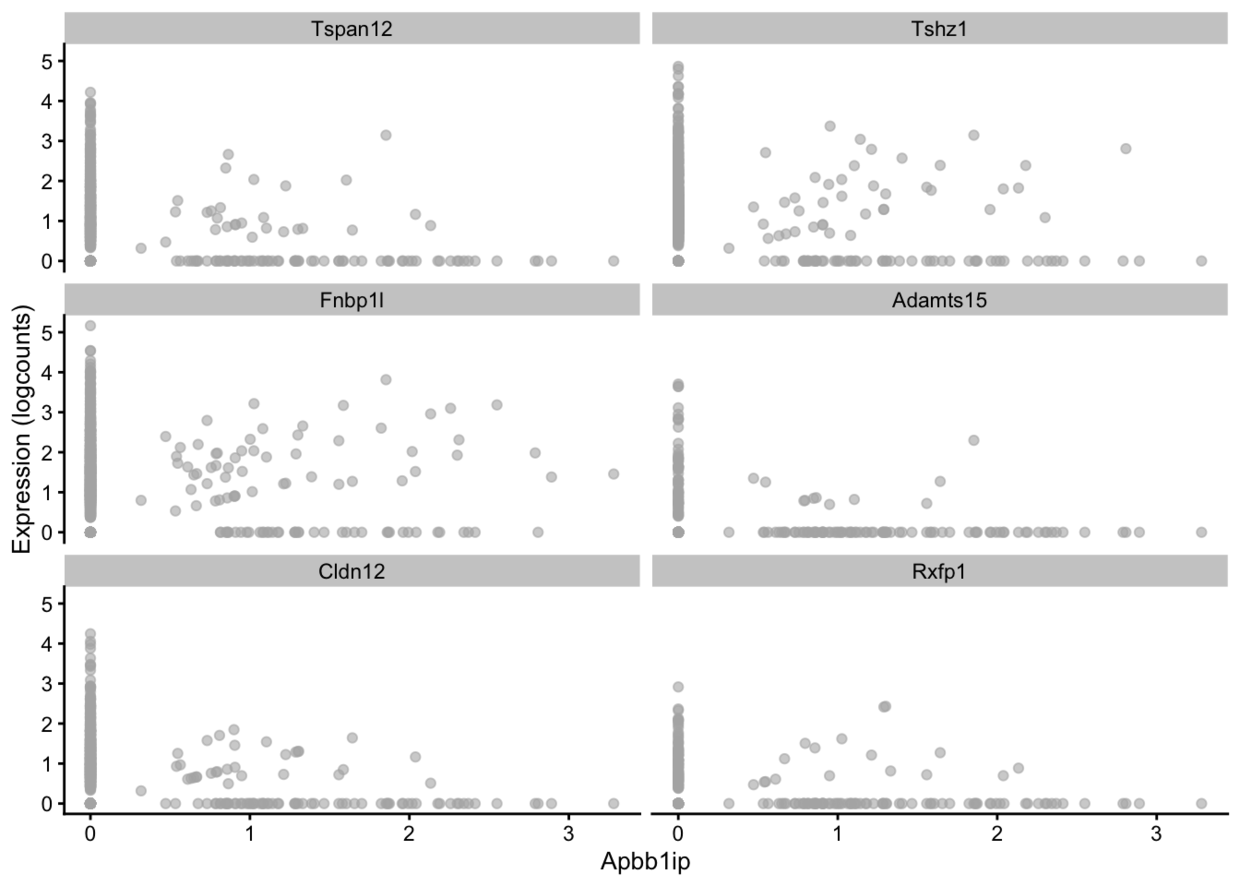
plotExpression(example_sce, rownames(example_sce)[1:6], x = "level1class")
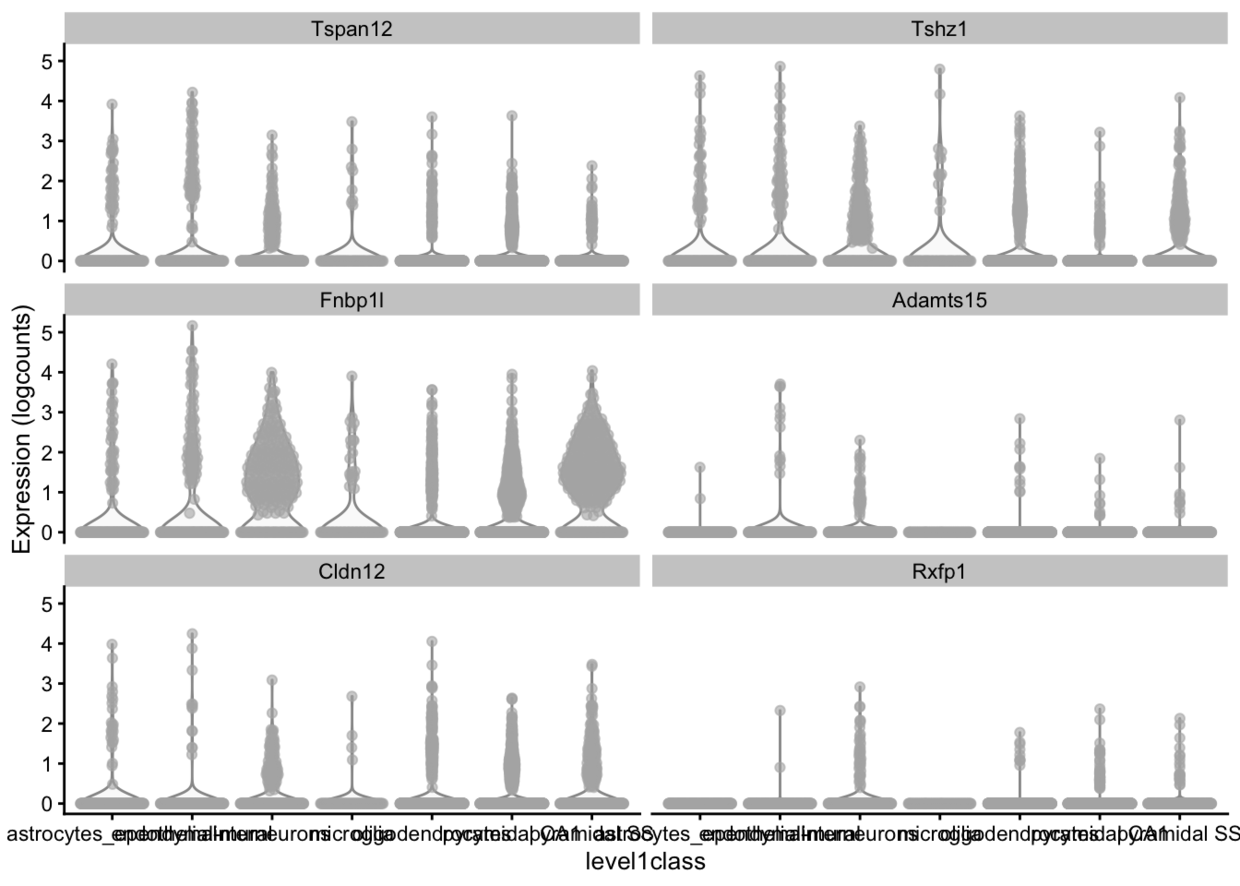
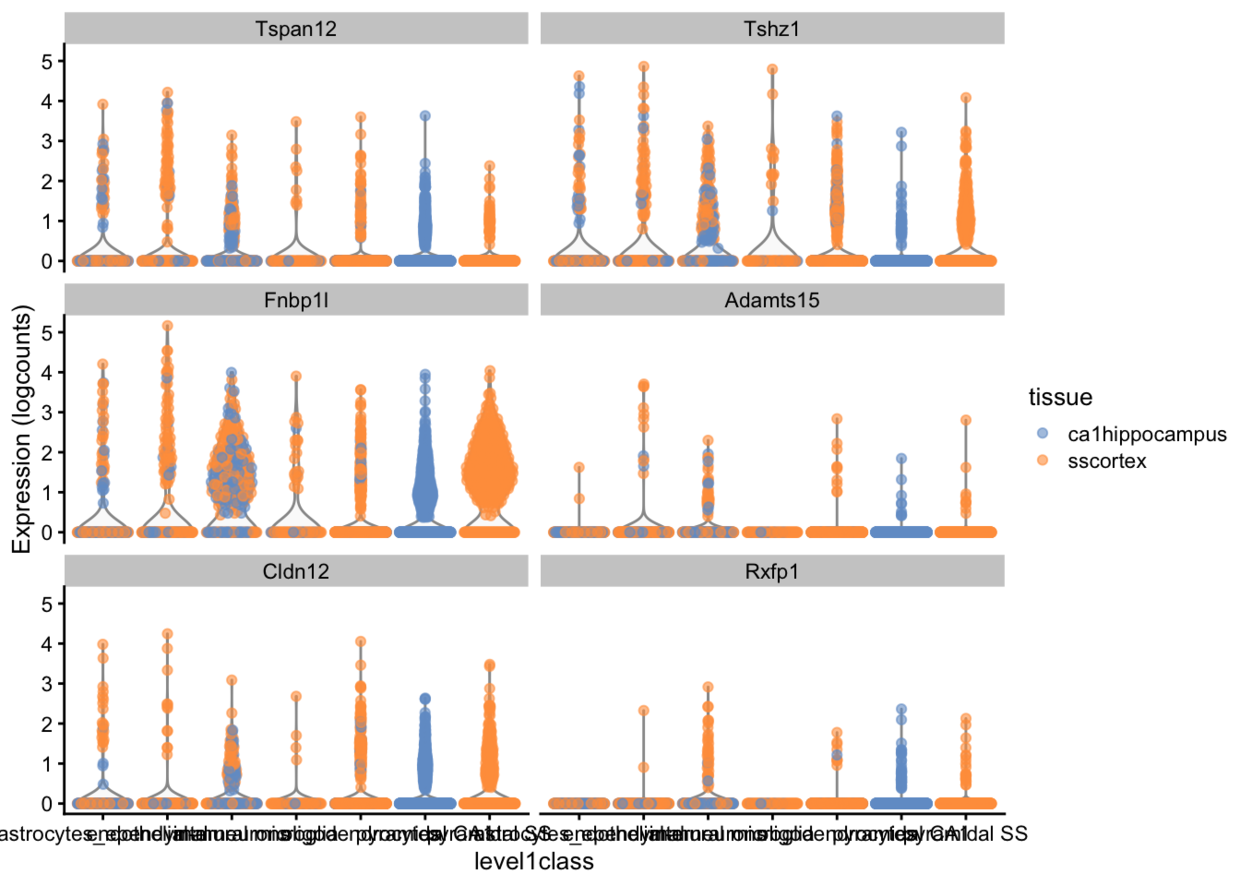
plotExpression(example_sce, rownames(example_sce)[1:6],x = "level1class", colour_by="tissue")
plotExpression(example_sce, rownames(example_sce)[1:6])
4.降维
默认情况下,runPCA()使用方差最高的前500个基因来计算PC1。默认计算50个主成分。可以用subset_row指定自选基因,用ncomponents指定要计算的主成分数。
PCA、t-SNE、UMAP三个方法
example_sce <- runPCA(example_sce)
str(reducedDim(example_sce, "PCA"))## num [1:3005, 1:50] -15.4 -15 -17.2 -16.9 -18.4 ...
## - attr(*, "dimnames")=List of 2
## ..$ : chr [1:3005] "1772071015_C02" "1772071017_G12" "1772071017_A05" "1772071014_B06" ...
## ..$ : chr [1:50] "PC1" "PC2" "PC3" "PC4" ...
## - attr(*, "varExplained")= num [1:50] 478 112.8 51.1 47 33.2 ...
## - attr(*, "percentVar")= num [1:50] 39.72 9.38 4.25 3.9 2.76 ...
## - attr(*, "rotation")= num [1:500, 1:50] 0.1471 0.1146 0.1084 0.0958 0.0953 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:500] "Plp1" "Trf" "Mal" "Apod" ...
## .. ..$ : chr [1:50] "PC1" "PC2" "PC3" "PC4" ...# set.seed(1000)
# example_sce < runTSNE(example_sce, perplexity=10)## 使用PCA结果的前10个维度来运行T-SNE
set.seed(1000)
example_sce <- runTSNE(example_sce, perplexity=50,dimred="PCA", n_dimred=10)example_sce <- runUMAP(example_sce)
降维的可视化。颜色可以根据colData里的已有注释,也可以根据某个基因的表达量来定义。下面代码里的Snap25和Mog都是基因名。
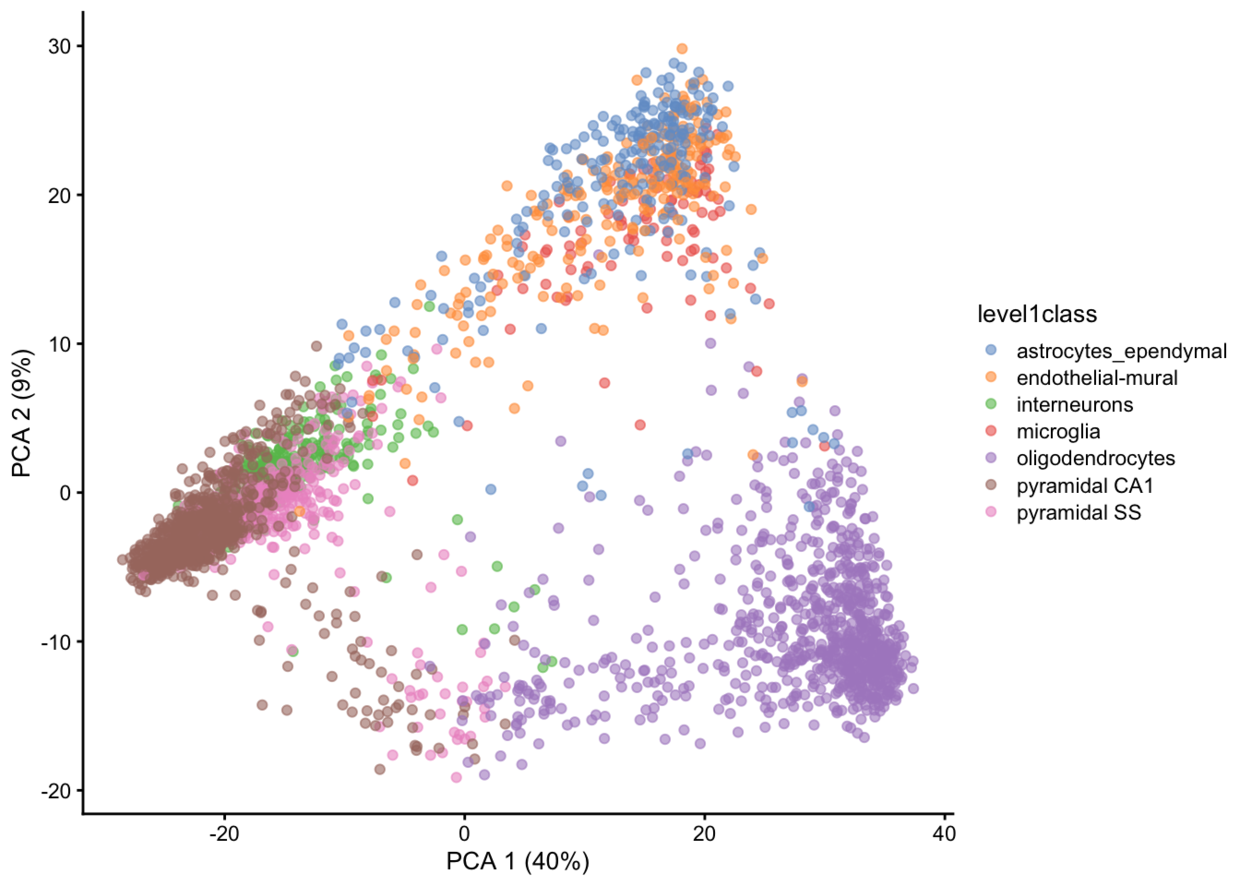
plotReducedDim(example_sce, dimred = "PCA", colour_by = "level1class")
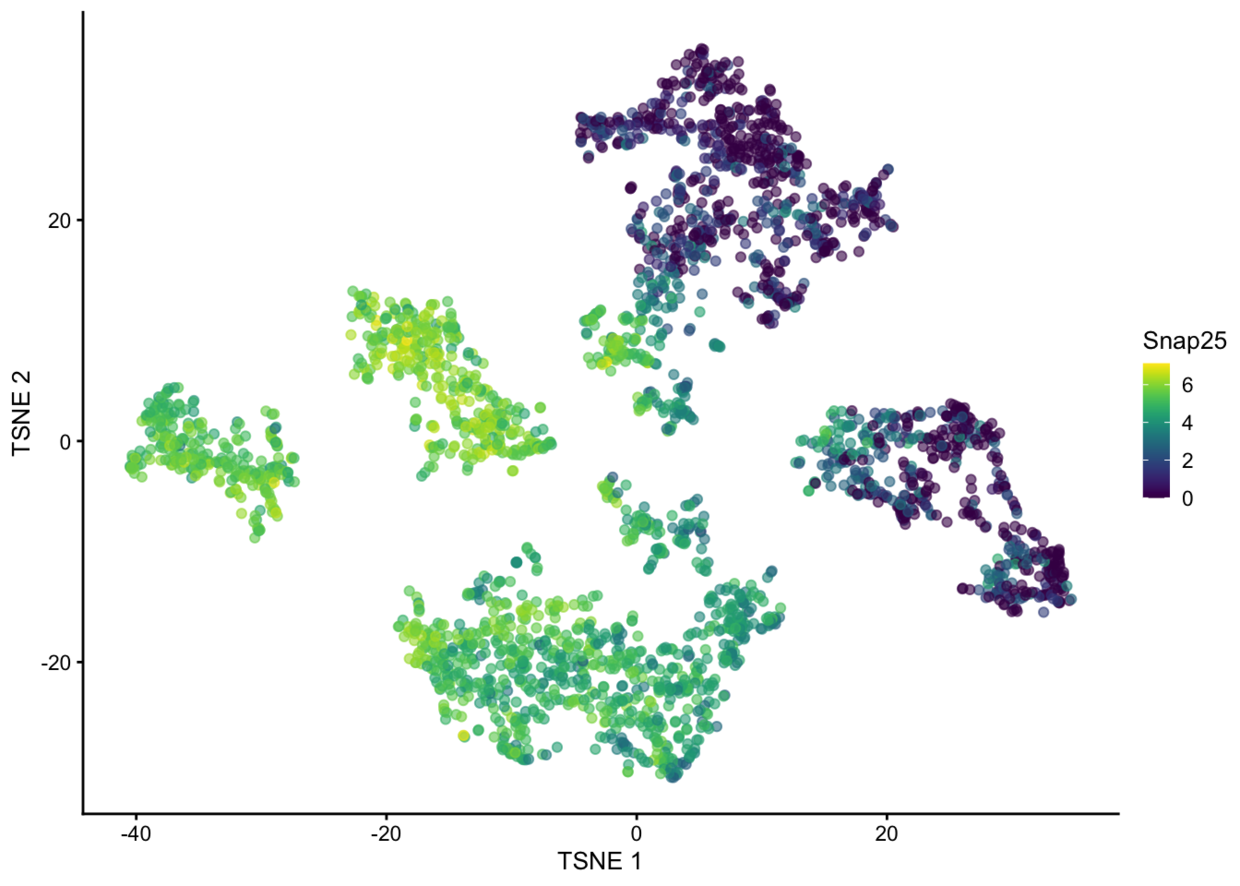
plotTSNE(example_sce, colour_by = "Snap25")
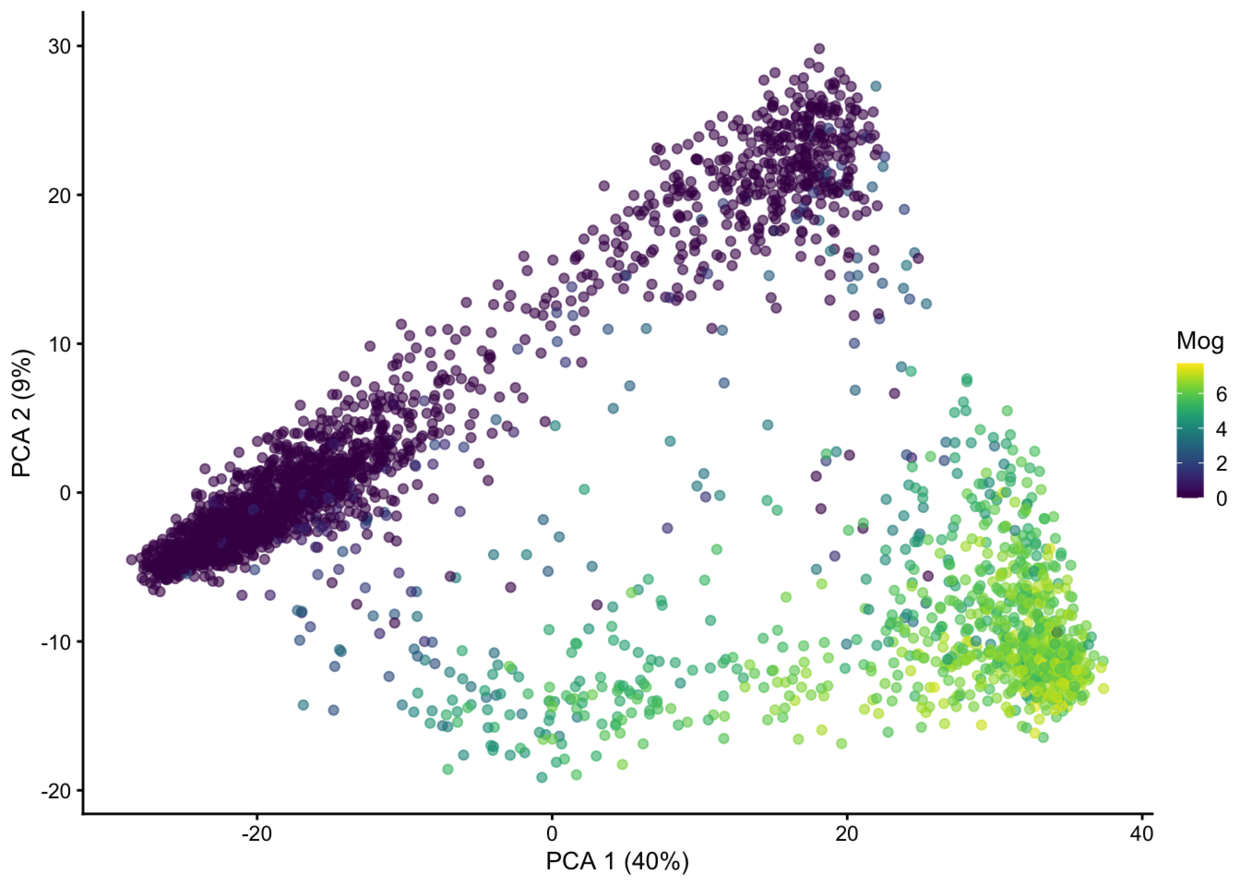
plotPCA(example_sce, colour_by="Mog")
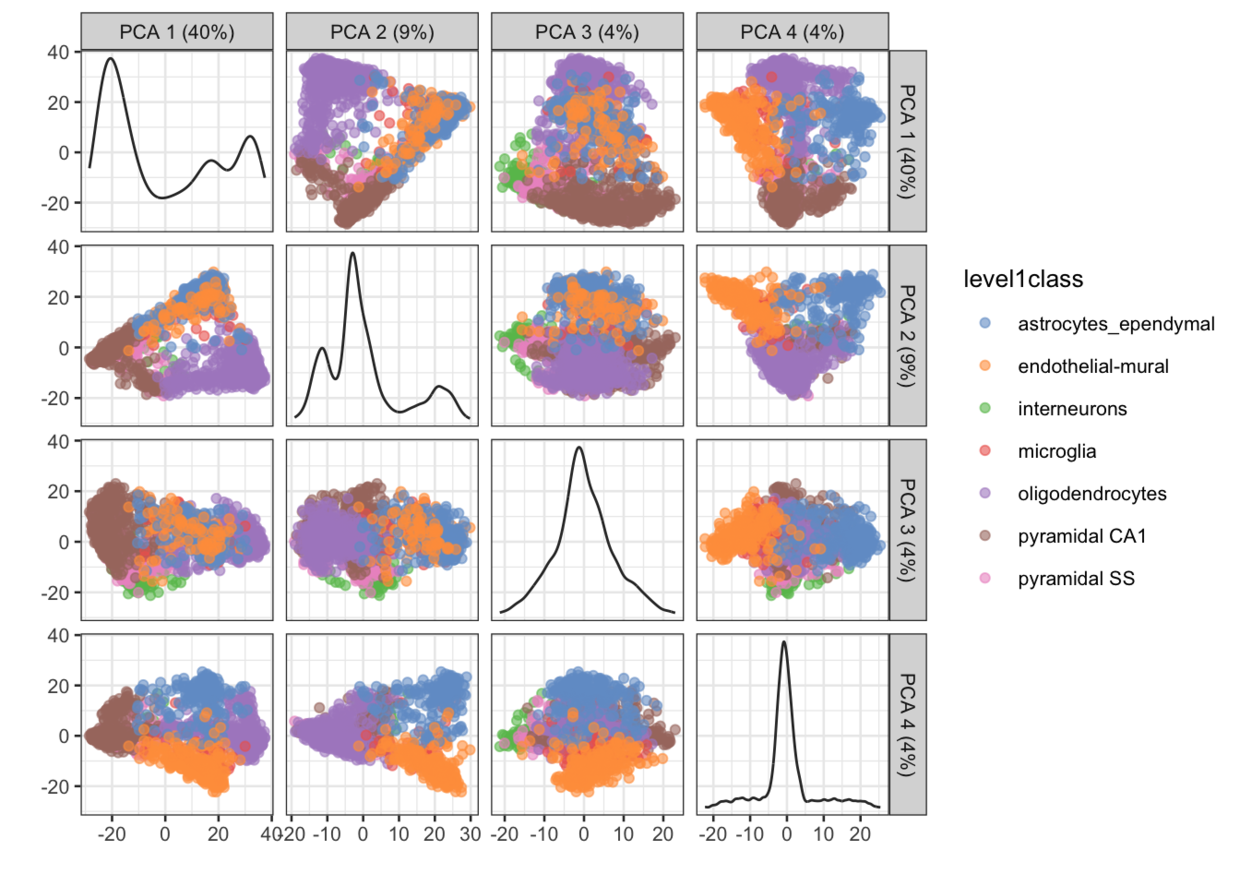
画多于两个维度的PCA图
plotPCA(example_sce, ncomponents = 4, colour_by = "level1class")
其他可视化
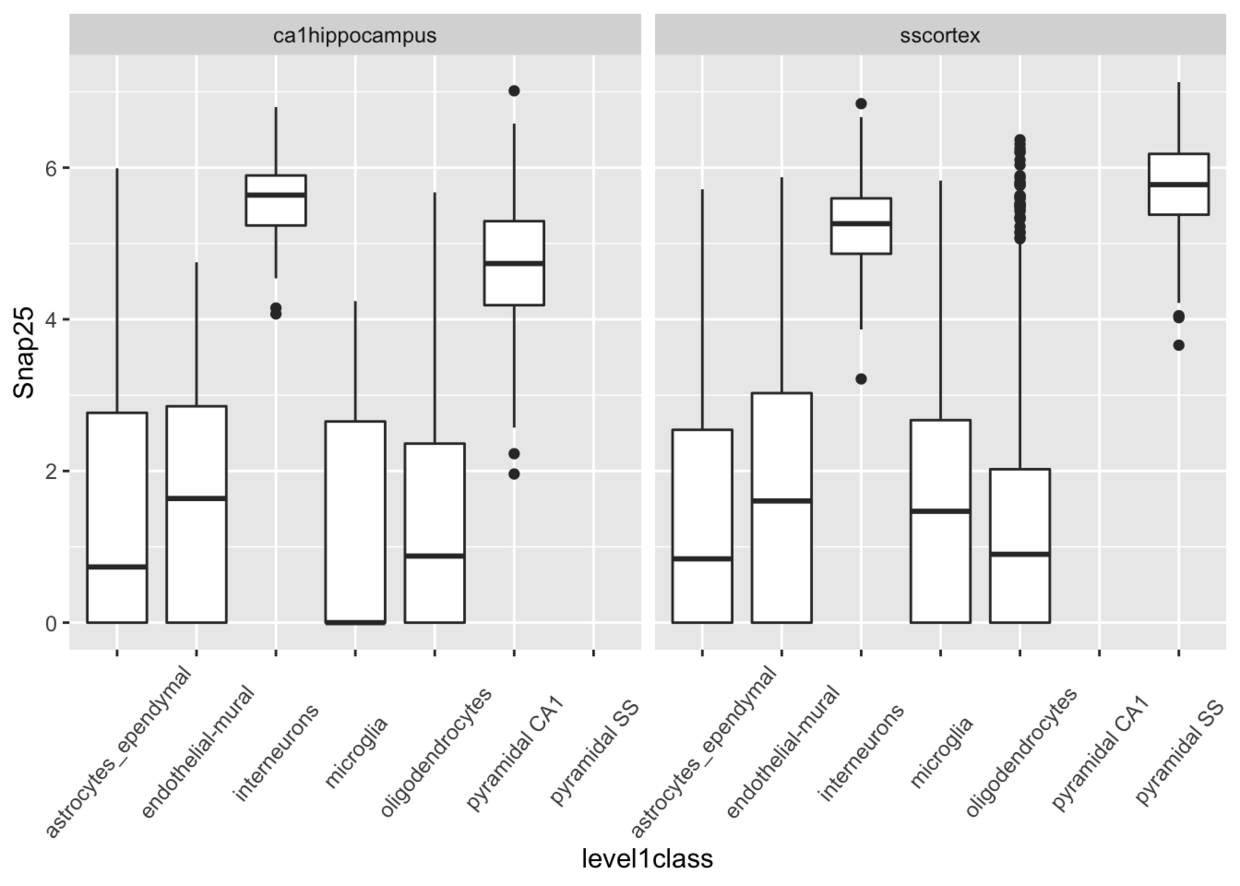
同一个基因在不同细胞间的表达量比较
ggcells(example_sce, mapping=aes(x=level1class, y=Snap25)) +geom_boxplot() +facet_wrap(~tissue)+ theme(axis.text.x = element_text(angle=50,vjust = 0.5))
ggcells(example_sce, mapping=aes(x=TSNE.1, y=TSNE.2, colour=Snap25)) +geom_point() +stat_density_2d() +facet_wrap(~tissue) +scale_colour_distiller(direction=1)
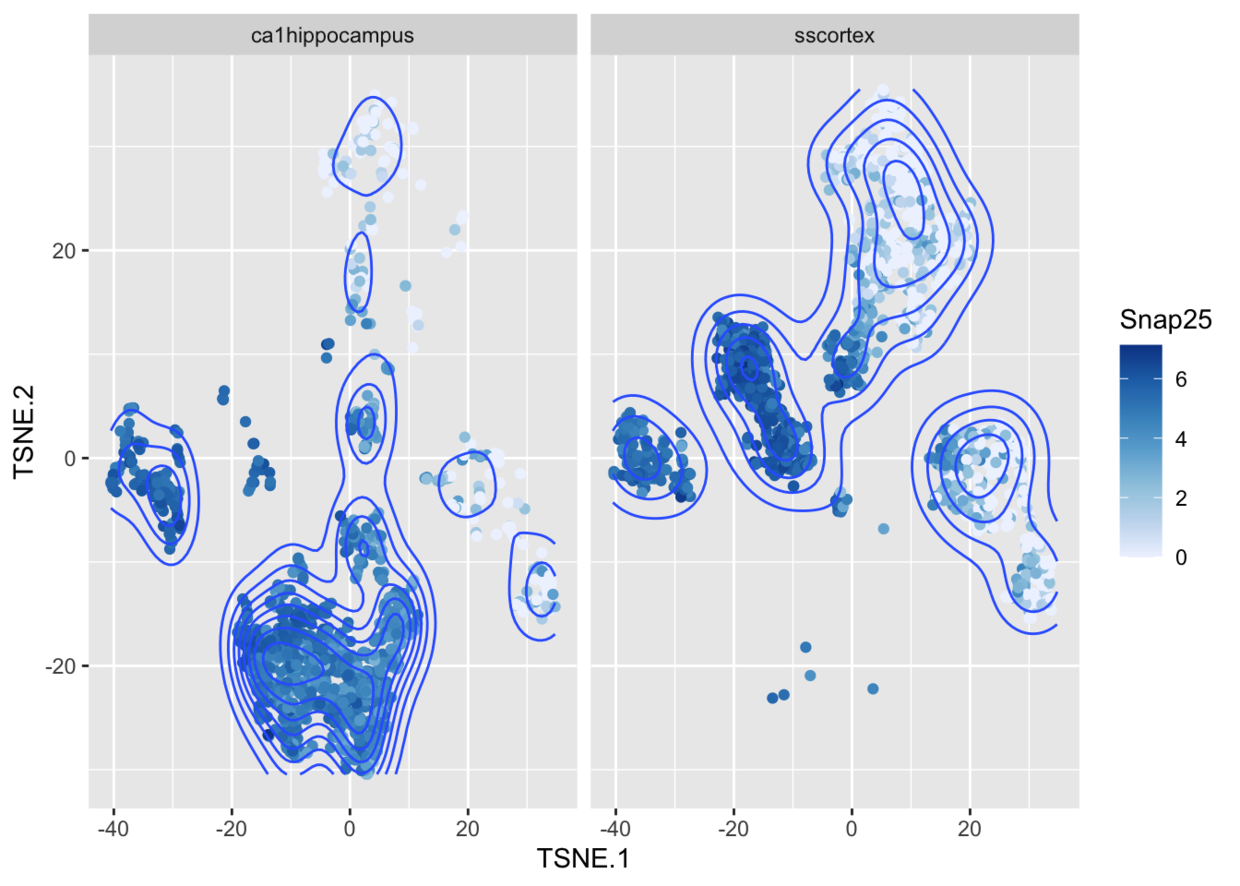
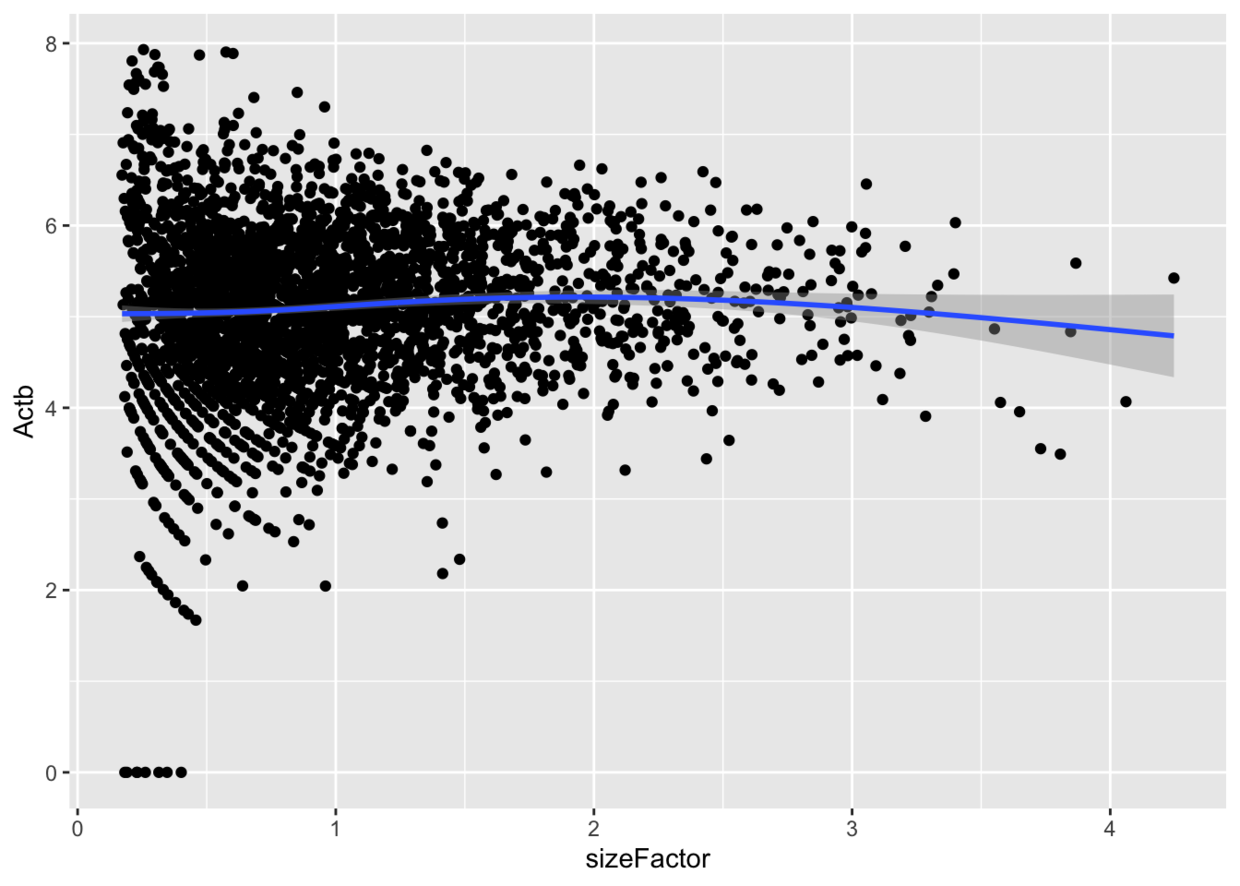
基因表达量与sizeFactor之间的关系
ggcells(example_sce, mapping=aes(x=sizeFactor, y=Actb)) +geom_point() +geom_smooth()
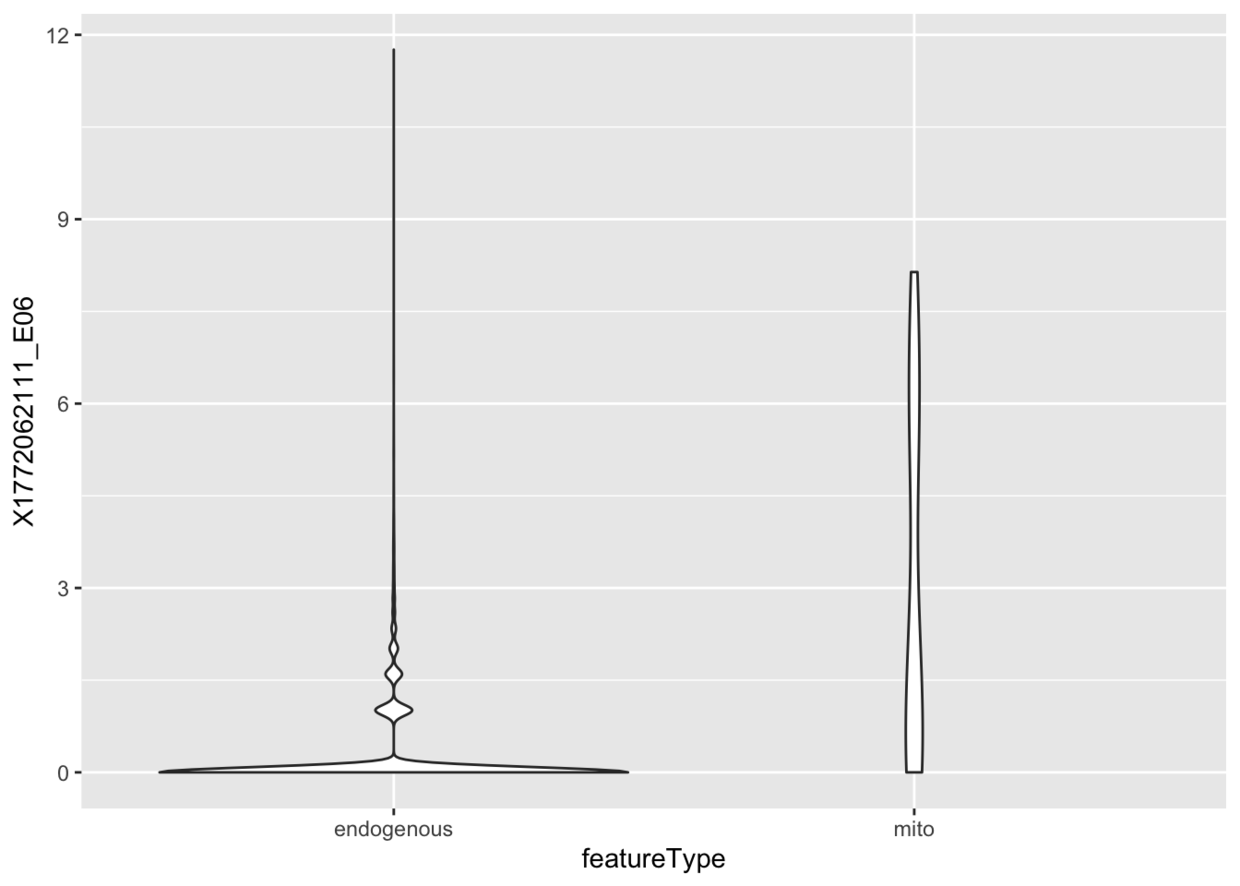
某一细胞的内源性基因与线粒体基因的表达量比较
colnames(example_sce) <- make.names(colnames(example_sce))
ggfeatures(example_sce, mapping=aes(x=featureType, y=X1772062111_E06)) +geom_violin()
本文来自互联网用户投稿,文章观点仅代表作者本人,不代表本站立场,不承担相关法律责任。如若转载,请注明出处。 如若内容造成侵权/违法违规/事实不符,请点击【内容举报】进行投诉反馈!