【图像分割】医学图像分割多目标分割(多分类)实践
文章目录
- 本文已更新到[【附源码】医学图像分割入门实践](https://blog.csdn.net/baidu_36511315/article/details/120902937)
- 1. 数据集
- 2. 数据预处理
- 3. 代码部分
- 3.1 训练集和验证集划分
- 3.2 数据加载和处理
- 3.2.1 数据变换
- 3.3 One-hot 工具函数
- 3.4 网络模型
- 3.5 模型权重初始化
- 3.6 损失函数
- 3.7 模型评价指标
- 3.8 训练
- 3.9 模型验证
- 3.10 实验结果
本文已更新到【附源码】医学图像分割入门实践
1. 数据集
来自ISICDM 2019 临床数据分析挑战赛的基于磁共振成像的膀胱内外壁分割与肿瘤检测数据集。
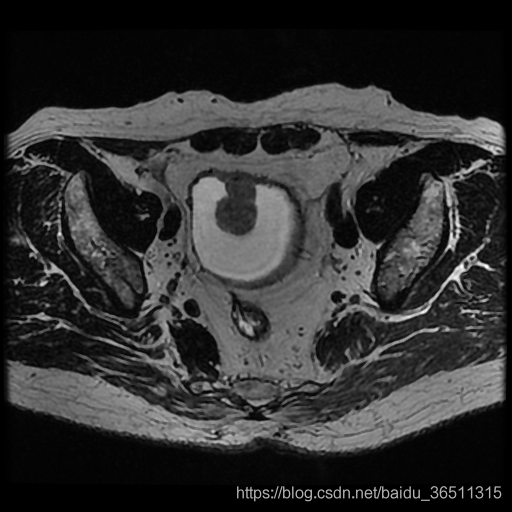

灰度值:灰色128为膀胱内外壁,白色255为肿瘤。
任务是要同时分割出膀胱内外壁和肿瘤部分,加上背景,最后构成一个三分类问题。
2. 数据预处理
数据预处理最重要的一步就是要对gt进行one-hot编码,如果对one-hot编码不太清楚可以看下这篇文章(数据预处理 One-hot 编码的两种实现方式)。
由于笔记本性能较差,为了代码能够在笔记本上跑起来。在对数据预处理的时候进行了缩放(scale)和中心裁剪(center crop)。原始数据大小为512,首先将数据缩放到256,再裁剪到128的大小。
3. 代码部分
3.1 训练集和验证集划分
按照训练集80%,验证集20%的策略进行重新分配数据集。直接运行当前文件进行数据重新划分,
仅供参考,当然这一部分代码可根据自己的需求随意设计。
# repartition_dataset.py
import os
import math
import randomdef partition_data(dataset_dir, ouput_root):"""Divide the raw data into training sets and validation sets:param dataset_dir: path root of dataset:param ouput_root: the root path to the output file:return:"""image_names = []mask_names = []val_size = 0.2train_names = []val_names = []for file in os.listdir(os.path.join(dataset_dir, "Images")):image_names.append(file)image_names.sort()for file in os.listdir(os.path.join(dataset_dir, "Labels")):mask_names.append(file)mask_names.sort()rawdata_size = len(image_names)random.seed(361)val_indices = random.sample(range(0, rawdata_size), math.floor(rawdata_size * val_size))train_indices = []for i in range(0, rawdata_size):if i not in val_indices:train_indices.append(i)with open(os.path.join(ouput_root, 'val.txt'), 'w') as f:for i in val_indices:val_names.append(image_names[i])f.write(image_names[i])f.write('\n')with open(os.path.join(ouput_root, 'train.txt'), 'w') as f:for i in train_indices:train_names.append(image_names[i])f.write(image_names[i])f.write('\n')train_names.sort(), val_names.sort()return train_names, val_namesif __name__ == '__main__':dataset_dir = '../media/LIBRARY/Datasets/Bladder/'output_root = '../media/LIBRARY/Datasets/Bladder/'train_names, val_names = partition_data(dataset_dir, output_root)print(len(train_names))print(train_names)print(len(val_names))print(val_names)
3.2 数据加载和处理
数据加载写一个专门的数据类来做就可以了,最核心的其实就是实现里面的__getitem__()方法。make_dataset方法用来加载数据的文件名,真正加载数据是在__getitem__()里面,在DataLoder的时候自动调用。
# baldder.py
import os
import cv2
import torch
import numpy as np
from PIL import Image
from torch.utils import data
from torchvision import transforms
from utils import helpers'''
128= bladder
255 = tumor
0 = background
'''
palette = [[0], [128], [255]]
num_classes = 3def make_dataset(root, mode):assert mode in ['train', 'val', 'test']items = []if mode == 'train':img_path = os.path.join(root, 'Images')mask_path = os.path.join(root, 'Labels')if 'Augdata' in root:data_list = os.listdir(os.path.join(root, 'Images'))else:data_list = [l.strip('\n') for l in open(os.path.join(root, 'train.txt')).readlines()]for it in data_list:item = (os.path.join(img_path, it), os.path.join(mask_path, it))items.append(item)elif mode == 'val':img_path = os.path.join(root, 'Images')mask_path = os.path.join(root, 'Labels')data_list = [l.strip('\n') for l in open(os.path.join(root, 'val.txt')).readlines()]for it in data_list:item = (os.path.join(img_path, it), os.path.join(mask_path, it))items.append(item)else:passreturn itemsclass Bladder(data.Dataset):def __init__(self, root, mode, joint_transform=None, center_crop=None, transform=None, target_transform=None):self.imgs = make_dataset(root, mode)self.palette = paletteself.mode = modeif len(self.imgs) == 0:raise RuntimeError('Found 0 images, please check the data set')self.mode = modeself.joint_transform = joint_transformself.center_crop = center_cropself.transform = transformself.target_transform = target_transformdef __getitem__(self, index):img_path, mask_path = self.imgs[index]img = Image.open(img_path)mask = Image.open(mask_path)if self.joint_transform is not None:img, mask = self.joint_transform(img, mask)if self.center_crop is not None:img, mask = self.center_crop(img, mask)img = np.array(img)mask = np.array(mask)# Image.open读取灰度图像时shape=(H, W) 而非(H, W, 1)# 因此先扩展出通道维度,以便在通道维度上进行one-hot映射img = np.expand_dims(img, axis=2)mask = np.expand_dims(mask, axis=2)mask = helpers.mask_to_onehot(mask, self.palette)# shape from (H, W, C) to (C, H, W)img = img.transpose([2, 0, 1])mask = mask.transpose([2, 0, 1])if self.transform is not None:img = self.transform(img)if self.target_transform is not None:mask = self.target_transform(mask)return img, maskdef __len__(self):return len(self.imgs)3.2.1 数据变换
# joint_transforms
import cv2
import math
import sys
import numbers
import random
from PIL import Image, ImageOps
import numpy as np
from skimage import measure
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
from utils import helpersclass Compose(object):def __init__(self, transforms):self.transforms = transformsdef __call__(self, img, mask):assert img.size == mask.sizefor t in self.transforms:img, mask = t(img, mask)return img, maskclass RandomCrop(object):def __init__(self, size, padding=0):if isinstance(size, numbers.Number):self.size = (int(size), int(size))else:self.size = sizeself.padding = paddingdef __call__(self, img, mask):if self.padding > 0:img = ImageOps.expand(img, border=self.padding, fill=0)mask = ImageOps.expand(mask, border=self.padding, fill=0)assert img.size == mask.sizew, h = img.sizeth, tw = self.sizeif w == tw and h == th:return img, maskif w < tw or h < th:return img.resize((tw, th), Image.BILINEAR), mask.resize((tw, th), Image.NEAREST)x1 = random.randint(0, w - tw)y1 = random.randint(0, h - th)return img.crop((x1, y1, x1 + tw, y1 + th)), mask.crop((x1, y1, x1 + tw, y1 + th))class CenterCrop(object):def __init__(self, size):if isinstance(size, numbers.Number):self.size = (int(size), int(size))else:self.size = sizedef __call__(self, img, mask):assert img.size == mask.sizew, h = img.sizeth, tw = self.sizex1 = int(math.ceil((w - tw) / 2.))y1 = int(math.ceil((h - th) / 2.))return img.crop((x1, y1, x1 + tw, y1 + th)), mask.crop((x1, y1, x1 + tw, y1 + th))class SingleCenterCrop(object):def __init__(self, size):if isinstance(size, numbers.Number):self.size = (int(size), int(size))else:self.size = sizedef __call__(self, img):w, h = img.sizeth, tw = self.sizex1 = int(math.ceil((w - tw) / 2.))y1 = int(math.ceil((h - th) / 2.))return img.crop((x1, y1, x1 + tw, y1 + th))class CenterCrop_npy(object):def __init__(self, size):self.size = sizedef __call__(self, img, mask):assert img.shape == mask.shapeif (self.size <= img.shape[1]) and (self.size <= img.shape[0]):x = math.ceil((img.shape[1] - self.size) / 2.)y = math.ceil((img.shape[0] - self.size) / 2.)if len(mask.shape) == 3:return img[y:y + self.size, x:x + self.size, :], mask[y:y + self.size, x:x + self.size, :]else:return img[y:y + self.size, x:x + self.size, :], mask[y:y + self.size, x:x + self.size]else:raise Exception('Crop shape (%d, %d) exceeds image dimensions (%d, %d)!' % (self.size, self.size, img.shape[0], img.shape[1]))class RandomHorizontallyFlip(object):def __call__(self, img, mask):if random.random() < 0.5:return img.transpose(Image.FLIP_LEFT_RIGHT), mask.transpose(Image.FLIP_LEFT_RIGHT)return img, maskclass Scale(object):def __init__(self, size):self.size = sizedef __call__(self, img, mask):assert img.size == mask.sizew, h = img.sizeif (w >= h and w == self.size) or (h >= w and h == self.size):return img, maskif w > h:ow = self.sizeoh = int(self.size * h / w)return img.resize((ow, oh), Image.BILINEAR), mask.resize((ow, oh), Image.NEAREST)else:oh = self.sizeow = int(self.size * w / h)return img.resize((ow, oh), Image.BILINEAR), mask.resize((ow, oh), Image.NEAREST)class RandomScaleCrop(object):def __init__(self, base_size, crop_size=0, scale_rate=0.95, fill=0):self.base_size = base_sizeself.crop_size = crop_sizeself.scale_rate = scale_rateself.fill = filldef __call__(self, im, gt):img = im.copy()mask = gt.copy()# random scale (short edge)short_size = random.randint(int(self.base_size * self.scale_rate), int(self.base_size * self.scale_rate))w, h = img.sizeif h > w:ow = short_sizeoh = int(1.0 * h * ow / w)else:oh = short_sizeow = int(1.0 * w * oh / h)img = img.resize((ow, oh), Image.BILINEAR)mask = mask.resize((ow, oh), Image.NEAREST)# pad cropif short_size < self.crop_size:padh = self.crop_size - oh if oh < self.crop_size else 0padw = self.crop_size - ow if ow < self.crop_size else 0img = ImageOps.expand(img, border=(0, 0, padw, padh), fill=0)mask = ImageOps.expand(mask, border=(0, 0, padw, padh), fill=0)w, h = img.sizex1 = random.randint(0, w - self.crop_size)y1 = random.randint(0, h - self.crop_size)img = img.crop((x1, y1, x1 + self.crop_size, y1 + self.crop_size))mask = mask.crop((x1, y1, x1 + self.crop_size, y1 + self.crop_size))return img, maskclass RandomRotate(object):def __init__(self, degree):self.degree = degreedef __call__(self, img, mask):rotate_degree = random.random() * 2 * self.degree - self.degreereturn img.rotate(rotate_degree, Image.BILINEAR), mask.rotate(rotate_degree, Image.NEAREST)# transforms.pyimport random
import numpy as np
import torch
from PIL import Image, ImageFilterclass DeNormalize(object):def __init__(self, mean, std):self.mean = meanself.std = stddef __call__(self, tensor):for t, m, s in zip(tensor, self.mean, self.std):t.mul_(s).add_(m)return tensorclass MaskToTensor(object):def __call__(self, img):return torch.from_numpy(np.array(img, dtype=np.float32))class NpyToTensor(object):def __call__(self, img):return torch.from_numpy(np.array(img, dtype=np.float32))# 不带归一化
class ImgToTensor(object):def __call__(self, img):img = torch.from_numpy(np.array(img))if isinstance(img, torch.ByteTensor):return img.float()3.3 One-hot 工具函数
# helpers.py
import os
import csv
import numpy as npdef mask_to_onehot(mask, palette):"""Converts a segmentation mask (H, W, C) to (H, W, K) where the last dim is a onehot encoding vector, C is usually 1 or 3, and K is the number of class."""semantic_map = []for colour in palette:equality = np.equal(mask, colour)class_map = np.all(equality, axis=-1)semantic_map.append(class_map)semantic_map = np.stack(semantic_map, axis=-1).astype(np.float32)return semantic_mapdef onehot_to_mask(mask, palette):"""Converts a mask (H, W, K) to (H, W, C)"""x = np.argmax(mask, axis=-1)colour_codes = np.array(palette)x = np.uint8(colour_codes[x.astype(np.uint8)])return x
3.4 网络模型
原始数据:shape = [N, 1, H, W]
GT: shape = [N, 3, H, W]
模型输出:shape = [N, 3, H, W]
(其中N为batch size的大小,H和W分别是图像的高和宽)
使用医学图像分割里面经典的U-Net网络。
# u_net.py
from torch import nn
from utils import initialize_weightsclass conv_block(nn.Module):def __init__(self, ch_in, ch_out):super(conv_block, self).__init__()self.conv = nn.Sequential(nn.Conv2d(ch_in, ch_out, kernel_size=3, stride=1, padding=1, bias=True),nn.BatchNorm2d(ch_out),nn.ReLU(inplace=True),nn.Conv2d(ch_out, ch_out, kernel_size=3, stride=1, padding=1, bias=True),nn.BatchNorm2d(ch_out),nn.ReLU(inplace=True))def forward(self, x):x = self.conv(x)return xclass up_conv(nn.Module):def __init__(self, ch_in, ch_out):super(up_conv, self).__init__()self.up = nn.Sequential(nn.Upsample(scale_factor=2),nn.Conv2d(ch_in, ch_out, kernel_size=3, stride=1, padding=1, bias=True),nn.BatchNorm2d(ch_out),nn.ReLU(inplace=True))def forward(self, x):x = self.up(x)return xclass U_Net(nn.Module):def __init__(self, img_ch=1, num_classes=3):super(U_Net, self).__init__()self.Maxpool = nn.MaxPool2d(kernel_size=2, stride=2)self.Conv1 = conv_block(ch_in=img_ch, ch_out=64)self.Conv2 = conv_block(ch_in=64, ch_out=128)self.Conv3 = conv_block(ch_in=128, ch_out=256)self.Conv4 = conv_block(ch_in=256, ch_out=512)self.Conv5 = conv_block(ch_in=512, ch_out=1024)self.Up5 = up_conv(ch_in=1024, ch_out=512)self.Up_conv5 = conv_block(ch_in=1024, ch_out=512)self.Up4 = up_conv(ch_in=512, ch_out=256)self.Up_conv4 = conv_block(ch_in=512, ch_out=256)self.Up3 = up_conv(ch_in=256, ch_out=128)self.Up_conv3 = conv_block(ch_in=256, ch_out=128)self.Up2 = up_conv(ch_in=128, ch_out=64)self.Up_conv2 = conv_block(ch_in=128, ch_out=64)self.Conv_1x1 = nn.Conv2d(64, num_classes, kernel_size=1, stride=1, padding=0)initialize_weights(self)def forward(self, x):# encoding pathx1 = self.Conv1(x)x2 = self.Maxpool(x1)x2 = self.Conv2(x2)x3 = self.Maxpool(x2)x3 = self.Conv3(x3)x4 = self.Maxpool(x3)x4 = self.Conv4(x4)x5 = self.Maxpool(x4)x5 = self.Conv5(x5)# decoding + concat pathd5 = self.Up5(x5)d5 = torch.cat((x4, d5), dim=1)d5 = self.Up_conv5(d5)d4 = self.Up4(d5)d4 = torch.cat((x3, d4), dim=1)d4 = self.Up_conv4(d4)d3 = self.Up3(d4)d3 = torch.cat((x2, d3), dim=1)d3 = self.Up_conv3(d3)d2 = self.Up2(d3)d2 = torch.cat((x1, d2), dim=1)d2 = self.Up_conv2(d2)d1 = self.Conv_1x1(d2)return d1
3.5 模型权重初始化
# utils.py
def initialize_weights(*models):for model in models:for module in model.modules():if isinstance(module, nn.Conv2d) or isinstance(module, nn.Linear):nn.init.kaiming_normal_(module.weight)if module.bias is not None:module.bias.data.zero_()elif isinstance(module, nn.BatchNorm2d):module.weight.data.fill_(1)module.bias.data.zero_()
3.6 损失函数
采用dice loss,实现思路可参考【Pytorch】 Dice系数与Dice Loss损失函数实现。
# loss.py
import torch.nn as nnfrom .metrics import *class SoftDiceLoss(_Loss):__name__ = 'dice_loss'def __init__(self, num_classes, activation=None, reduction='mean'):super(SoftDiceLoss, self).__init__()self.activation = activationself.num_classes = num_classesdef forward(self, y_pred, y_true):class_dice = []for i in range(1, self.num_classes):class_dice.append(diceCoeff(y_pred[:, i:i + 1, :], y_true[:, i:i + 1, :], activation=self.activation))mean_dice = sum(class_dice) / len(class_dice)return 1 - mean_dice
3.7 模型评价指标
Dice 系数。
# metircs.pyimport torch
import torch.nn as nn
import numpy as npdef diceCoeff(pred, gt, eps=1e-5, activation='sigmoid'):r""" computational formula:dice = (2 * (pred ∩ gt)) / (pred ∪ gt)"""if activation is None or activation == "none":activation_fn = lambda x: xelif activation == "sigmoid":activation_fn = nn.Sigmoid()elif activation == "softmax2d":activation_fn = nn.Softmax2d()else:raise NotImplementedError("Activation implemented for sigmoid and softmax2d")pred = activation_fn(pred)N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)intersection = (pred_flat * gt_flat).sum(1)unionset = pred_flat.sum(1) + gt_flat.sum(1)loss = (2 * intersection + eps) / (unionset + eps)return loss.sum() / Ndef diceCoeffv2(pred, gt, eps=1e-5, activation='sigmoid'):r""" computational formula:dice = (2 * tp) / (2 * tp + fp + fn)"""if activation is None or activation == "none":activation_fn = lambda x: xelif activation == "sigmoid":activation_fn = nn.Sigmoid()elif activation == "softmax2d":activation_fn = nn.Softmax2d()else:raise NotImplementedError("Activation implemented for sigmoid and softmax2d")pred = activation_fn(pred)N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)tp = torch.sum(gt_flat * pred_flat, dim=1)fp = torch.sum(pred_flat, dim=1) - tpfn = torch.sum(gt_flat, dim=1) - tploss = (2 * tp + eps) / (2 * tp + fp + fn + eps)return loss.sum() / N
3.8 训练
# train.py
import time
import os
from torch import optim
from torch.utils.data import DataLoader
from tensorboardX import SummaryWriter# from datasets import bladder
from utils.loss import *
from utils import tools
from utils.metrics import diceCoeffv2
import utils.joint_transforms as joint_transforms
import utils.transforms as extended_transforms
from networks.u_net import *crop_size = 128
batch_size = 2
n_epoch = 10
model_name = 'U_Net_'
loss_name = 'dice_'
times = 'no1_'
extra_description = ''
writer = SummaryWriter(os.path.join('../../log/bladder_trainlog', 'bladder_exp', model_name+loss_name+times+extra_description))def main():net = U_Net(img_ch=1, num_classes=3).cuda()train_joint_transform = joint_transforms.Compose([joint_transforms.Scale(256),# joint_transforms.RandomRotate(10),# joint_transforms.RandomHorizontallyFlip()])center_crop = joint_transforms.CenterCrop(crop_size)train_input_transform = extended_transforms.ImgToTensor()target_transform = extended_transforms.MaskToTensor()train_set = bladder.Bladder('../../media/LIBRARY/Datasets/Bladder', 'train',joint_transform=train_joint_transform, center_crop=center_crop,transform=train_input_transform, target_transform=target_transform)train_loader = DataLoader(train_set, batch_size=batch_size, shuffle=True)if loss_name == 'dice_':criterion = SoftDiceLoss(activation='sigmoid').cuda()elif loss_name == 'bce_':criterion = nn.BCEWithLogitsLoss().cuda()elif loss_name == 'wbce_':criterion = WeightedBCELossWithSigmoid().cuda()elif loss_name == 'er_':criterion = EdgeRefinementLoss().cuda()optimizer = optim.Adam(net.parameters(), lr=1e-4)train(train_loader, net, criterion, optimizer, n_epoch, 0)def train(train_loader, net, criterion, optimizer, num_epoches , iters):for epoch in range(1, num_epoches + 1):st = time.time()b_dice = 0.0t_dice = 0.0d_len = 0for inputs, mask in train_loader:X = inputs.cuda()y = mask.cuda()optimizer.zero_grad()output = net(X)loss = criterion(output, y)# CrossEntropyLoss# loss = criterion(output, torch.argmax(y, dim=1))output = torch.sigmoid(output)output[output < 0.5] = 0output[output > 0.5] = 1bladder_dice = diceCoeffv2(output[:, 0:1, :], y[:, 0:1, :], activation=None).cpu().item()tumor_dice = diceCoeffv2(output[:, 1:2, :], y[:, 1:2, :], activation=None).cpu().item()mean_dice = (bladder_dice + tumor_dice) / 2d_len += 1b_dice += bladder_dicet_dice += tumor_diceloss.backward()optimizer.step()iters += batch_sizestring_print = "Epoch = %d iters = %d Current_Loss = %.4f Mean Dice=%.4f Bladder Dice=%.4f Tumor Dice=%.4f Time = %.2f"\% (epoch, iters, loss.item(), mean_dice,bladder_dice, tumor_dice, time.time() - st)tools.log(string_print)st = time.time()writer.add_scalar('train_main_loss', loss.item(), iters)b_dice = b_dice / d_lent_dice = t_dice / d_lenm_dice = (b_dice + t_dice) / 2print('Epoch {}/{},Train Mean Dice {:.4}, Bladder Dice {:.4}, Tumor Dice {:.4}'.format(epoch, num_epoches, m_dice, b_dice, t_dice))if epoch == num_epoches:torch.save(net, '../../checkpoint/exp/{}.pth'.format(model_name + loss_name + times + extra_description))writer.close()if __name__ == '__main__':main()
3.9 模型验证
# validate.py
import os
import cv2
from PIL import Image
import utils.joint_transforms as joint_transforms
from torch.utils.data import DataLoader
from tensorboardX import SummaryWriterfrom utils import helpers
import utils.transforms as extended_transforms
from utils.metrics import *
from datasets import bladder
from utils.loss import *
import trainLOSS = False
# numpy 高维数组打印不显示...
np.set_printoptions(threshold=9999999)
batch_size = 1val_input_transform = extended_transforms.ImgToTensor()
center_crop = joint_transforms.Compose([joint_transforms.Scale(256),joint_transforms.CenterCrop(128)]
)target_transform = extended_transforms.MaskToTensor()
val_set = bladder.Bladder('../../media/LIBRARY/Datasets/Bladder/', 'val',transform=val_input_transform, center_crop=center_crop,target_transform=target_transform)
val_loader = DataLoader(val_set, batch_size=batch_size, shuffle=False)# 验证用的模型名称
model_name = train.model_name
loss_name = train.loss_name
times = train.times
extra_description = train.extra_description
model = torch.load("../../checkpoint/exp/{}.pth".format(model_name + loss_name + times + extra_description))
# model = torch.load("../../checkpoint/exp/{}.pth".format('U_Net_bce_no1_'))
model.eval()
if LOSS:writer = SummaryWriter(os.path.join('../../log/vallog', 'bladder_exp', model_name+loss_name+times+extra_description))if loss_name == 'dice_':criterion = SoftDiceLoss(activation='sigmoid').cuda()
elif loss_name == 'bce_':criterion = nn.BCEWithLogitsLoss().cuda()
elif loss_name == 'wbce_':criterion = WeightedBCELossWithSigmoid().cuda()
elif loss_name == 'er_':criterion = EdgeRefinementLoss().cuda()def val(model):imname = '2-IM131'# imname = '2-IM107'img = Image.open('D:\\Learning\\datasets\\基于磁共振成像的膀胱内外壁分割与肿瘤检测\\Images\\{}.png'.format(imname))mask = Image.open('D:\\Learning\\datasets\\基于磁共振成像的膀胱内外壁分割与肿瘤检测\\Labels\\{}.png'.format(imname))img, mask = center_crop(img, mask)img = np.asarray(img)img = np.expand_dims(img, axis=2)mri = imgmask = np.asarray(mask)mask = np.expand_dims(mask, axis=2)gt = np.float32(helpers.mask_to_onehot(mask, bladder.palette))# 用来看gt的像素值gt_showval = gtgt = np.expand_dims(gt, axis=3)gt = gt.transpose([3, 2, 0, 1])gt = torch.from_numpy(gt)img = img.transpose([2, 0, 1])img = np.expand_dims(img, axis=3)img = img.transpose([3, 0, 1, 2])img = val_input_transform(img)img = img.cuda()model = model.cuda()pred = model(img)pred = torch.sigmoid(pred)pred[pred < 0.5] = 0pred[pred > 0.5] = 1bladder_dice = diceCoeffv2(pred[:, 0:1, :], gt.cuda()[:, 0:1, :], activation=None)tumor_dice = diceCoeffv2(pred[:, 1:2, :], gt.cuda()[:, 1:2, :], activation=None)mean_dice = (bladder_dice + tumor_dice) / 2acc = accuracy(pred, gt.cuda())p = precision(pred, gt.cuda())r = recall(pred, gt.cuda())print('mean_dice={:.4}, bladder_dice={:.4}, tumor_dice={:.4}, acc={:.4}, p={:.4}, r={:.4}'.format(mean_dice.item(), bladder_dice.item(), tumor_dice.item(),acc.item(), p.item(), r.item()))pred = pred.cpu().detach().numpy()[0].transpose([1, 2, 0])# 用来看预测的像素值pred_showval = predpred = helpers.onehot_to_mask(pred, bladder.palette)# np.uint8()反归一化到[0, 255]imgs = np.uint8(np.hstack([mri, pred, mask]))cv2.imshow("mri pred gt", imgs)cv2.waitKey(0)def auto_val(model):# 效果展示图片数iters = 0SIZES = 8imgs = []preds = []gts = []dices = 0tumor_dices = 0bladder_dices = 0for i, (img, mask) in enumerate(val_loader):im = imgimg = img.cuda()model = model.cuda()pred = model(img)if LOSS:loss = criterion(pred, mask.cuda()).item()pred = torch.sigmoid(pred)pred = pred.cpu().detach()iters += batch_sizepred[pred < 0.5] = 0pred[pred > 0.5] = 1bladder_dice = diceCoeff(pred[:, 0:1, :], mask[:, 0:1, :], activation=None)tumor_dice = diceCoeff(pred[:, 1:2, :], mask[:, 1:2, :], activation=None)mean_dice = (bladder_dice + tumor_dice) / 2dices += mean_dicetumor_dices += tumor_dicebladder_dices += bladder_diceacc = accuracy(pred, mask)p = precision(pred, mask)r = recall(pred, mask)print('mean_dice={:.4}, bladder_dice={:.4}, tumor_dice={:.4}, acc={:.4}, p={:.4}, r={:.4}'.format(mean_dice.item(), bladder_dice.item(), tumor_dice.item(),acc, p, r))gt = mask.numpy()[0].transpose([1, 2, 0])gt = helpers.onehot_to_mask(gt, bladder.palette)pred = pred.cpu().detach().numpy()[0].transpose([1, 2, 0])pred = helpers.onehot_to_mask(pred, bladder.palette)im = im[0].numpy().transpose([1, 2, 0])if LOSS:writer.add_scalar('val_main_loss', loss, iters)if len(imgs) < SIZES:imgs.append(im * 255)preds.append(pred)gts.append(gt)val_mean_dice = dices / (len(val_loader) / batch_size)val_tumor_dice = tumor_dices / (len(val_loader) / batch_size)val_bladder_dice = bladder_dices / (len(val_loader) / batch_size)print('Val Mean Dice = {:.4}, Val Bladder Dice = {:.4}, Val Tumor Dice = {:.4}'.format(val_mean_dice, val_bladder_dice, val_tumor_dice))imgs = np.hstack([*imgs])preds = np.hstack([*preds])gts = np.hstack([*gts])show_res = np.vstack(np.uint8([imgs, preds, gts]))cv2.imshow("top is mri , middle is pred, bottom is gt", show_res)cv2.waitKey(0)if __name__ == '__main__':# val(model)auto_val(model)
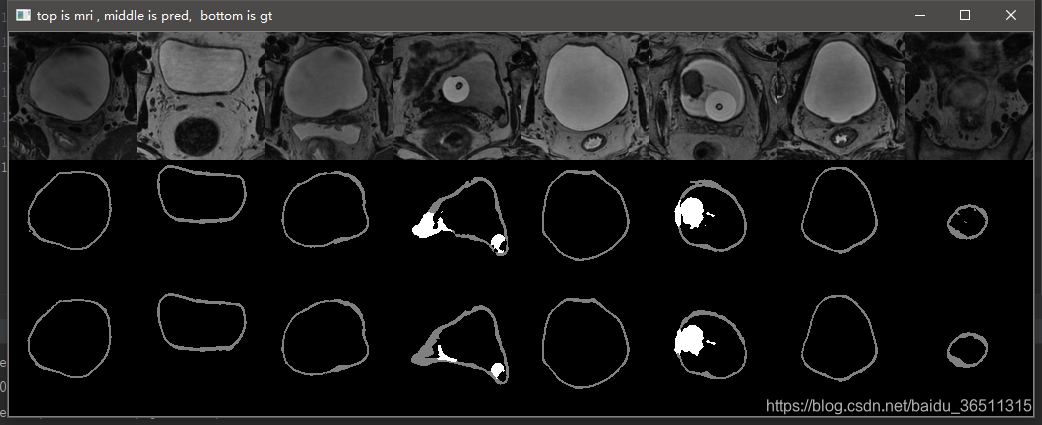
3.10 实验结果
这是笔记本跑了10个epoch的结果,仅仅是测试代码有没有问题。从结果可以看到,代码目前应该是没有问题的,后期只需调参数再训练提升效果即可。
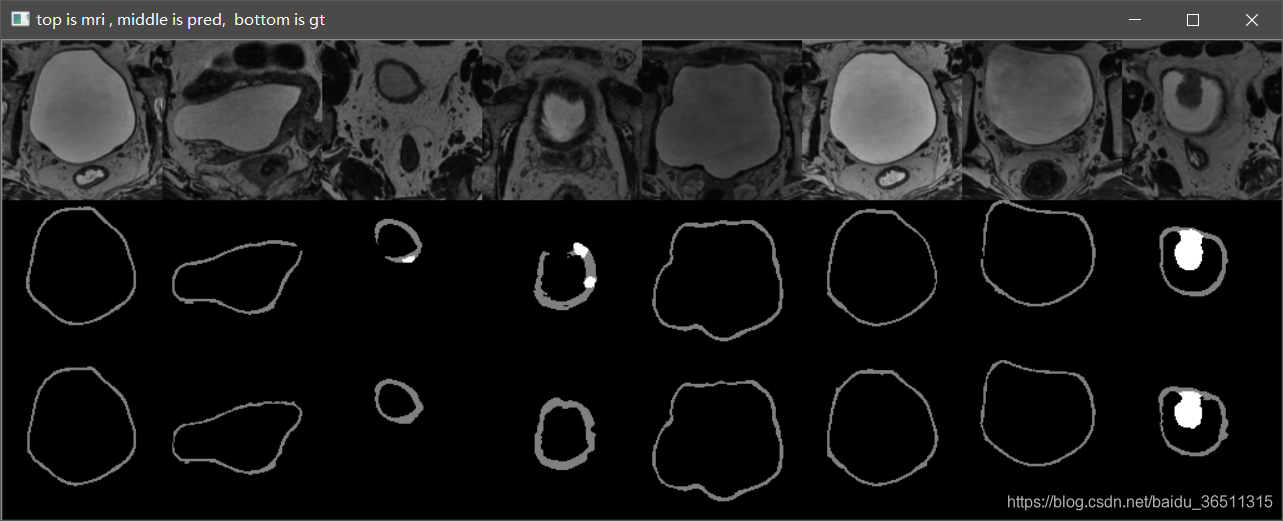
GTX2080TI 跑120个epoch的测试效果:
Val Mean Dice = 0.9051, Val Bladder Dice = 0.9012, Val Tumor Dice = 0.9091
- 2020/10/19更新:评价指标代码
import torch
import torch.nn as nn
import numpy as np"""
True Positive (真正, TP)预测为正的正样本
True Negative(真负 , TN)预测为负的负样本
False Positive (假正, FP)预测为正的负样本
False Negative(假负 , FN)预测为负的正样本
"""def diceCoeff(pred, gt, smooth=1e-5, ):r""" computational formula:dice = (2 * (pred ∩ gt)) / (pred ∪ gt)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)intersection = (pred_flat * gt_flat).sum(1)unionset = pred_flat.sum(1) + gt_flat.sum(1)score = (2 * intersection + smooth) / (unionset + smooth)return score.sum() / Ndef diceCoeffv2(pred, gt, eps=1e-5):r""" computational formula:dice = (2 * tp) / (2 * tp + fp + fn)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)tp = torch.sum(gt_flat * pred_flat, dim=1)fp = torch.sum(pred_flat, dim=1) - tpfn = torch.sum(gt_flat, dim=1) - tpscore = (2 * tp + eps) / (2 * tp + fp + fn + eps)return score.sum() / Ndef diceCoeffv3(pred, gt, eps=1e-5):r""" computational formula:dice = (2 * tp) / (2 * tp + fp + fn)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)tp = torch.sum((pred_flat != 0) * (gt_flat != 0), dim=1)fp = torch.sum((pred_flat != 0) * (gt_flat == 0), dim=1)fn = torch.sum((pred_flat == 0) * (gt_flat != 0), dim=1)# 转为float,以防long类型之间相除结果为0score = (2 * tp + eps).float() / (2 * tp + fp + fn + eps).float()return score.sum() / Ndef jaccard(pred, gt):"""TP / (TP + FP + FN)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)tp = torch.sum((pred_flat != 0) * (gt_flat != 0))fp = torch.sum((pred_flat != 0) * (gt_flat == 0))fn = torch.sum((pred_flat == 0) * (gt_flat != 0))score = tp.float() / (tp + fp + fn).float()return score.sum() / Ndef tversky(pred, gt, eps=1e-5, alpha=0.7):"""TP / (TP + (1-alpha) * FP + alpha * FN)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)tp = torch.sum(gt_flat * pred_flat, dim=1)fp = torch.sum(pred_flat, dim=1) - tpfn = torch.sum(gt_flat, dim=1) - tpscore = (tp + eps) / (tp + (1-alpha) * fp + alpha*fn + eps)return score.sum() / Ndef accuracy(pred, gt):"""(TP + TN) / (TP + FP + FN + TN)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)tp = torch.sum((pred_flat != 0) * (gt_flat != 0))fp = torch.sum((pred_flat != 0) * (gt_flat == 0))tn = torch.sum((pred_flat == 0) * (gt_flat == 0))fn = torch.sum((pred_flat == 0) * (gt_flat != 0))score = (tp + tn).float() / (tp + fp + tn + fn).float()return score.sum() / Ndef precision(pred, gt):"""TP / (TP + FP)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)tp = torch.sum((pred_flat != 0) * (gt_flat != 0))fp = torch.sum((pred_flat != 0) * (gt_flat == 0))score = tp.float() / (tp + fp).float()return score.sum() / Ndef sensitivity(pred, gt):"""TP / (TP + FN)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)tp = torch.sum((pred_flat != 0) * (gt_flat != 0))fn = torch.sum((pred_flat == 0) * (gt_flat != 0))score = tp.float() / (tp + fn).float()return score.sum() / Ndef specificity(pred, gt):"""TN / (TN + FP)"""N = gt.size(0)pred_flat = pred.view(N, -1)gt_flat = gt.view(N, -1)fp = torch.sum((pred_flat != 0) * (gt_flat == 0))tn = torch.sum((pred_flat == 0) * (gt_flat == 0))score = tn.float() / (fp + tn).float()return score.sum() / Ndef recall(pred, gt):return sensitivity(pred, gt)if __name__ == '__main__':# shape = torch.Size([2, 3, 4, 4])# 模拟batch_size = 2'''1 0 0= bladder0 1 0 = tumor0 0 1= background '''pred = torch.Tensor([[[[0, 1, 0, 0],[1, 0, 0, 1],[1, 0, 0, 1],[0, 1, 1, 0]],[[0, 0, 0, 0],[0, 0, 0, 0],[0, 1, 1, 0],[0, 0, 0, 0]],[[1, 0, 1, 1],[0, 1, 1, 0],[0, 0, 0, 0],[1, 0, 0, 1]]],[[[0, 1, 0, 0],[1, 0, 0, 1],[1, 0, 0, 1],[0, 1, 1, 0]],[[0, 0, 0, 0],[0, 0, 0, 0],[0, 1, 1, 0],[0, 0, 0, 0]],[[1, 0, 1, 1],[0, 1, 1, 0],[0, 0, 0, 0],[1, 0, 0, 1]]]])gt = torch.Tensor([[[[0, 1, 1, 0],[1, 0, 0, 1],[1, 0, 0, 1],[0, 1, 1, 0]],[[0, 0, 0, 0],[0, 0, 0, 0],[0, 1, 1, 0],[0, 0, 0, 0]],[[1, 0, 0, 1],[0, 1, 1, 0],[0, 0, 0, 0],[1, 0, 0, 1]]],[[[0, 1, 1, 0],[1, 0, 0, 1],[1, 0, 0, 1],[0, 1, 1, 0]],[[0, 0, 0, 0],[0, 0, 0, 0],[0, 1, 1, 0],[0, 0, 0, 0]],[[1, 0, 0, 1],[0, 1, 1, 0],[0, 0, 0, 0],[1, 0, 0, 1]]]])dice1 = diceCoeff(pred[:, 0:1, :], gt[:, 0:1, :], activation=None)dice2 = jaccard(pred[:, 0:1, :], gt[:, 0:1, :], activation=None)dice3 = diceCoeffv3(pred[:, 0:1, :], gt[:, 0:1, :], activation=None)print(dice1, dice2, dice3)
tips:有些指标的代码我还没测试过,目前dice用的比较多,有问题的话还望谅解^^
- 2021/5/9更新
好多小伙伴私信要github源码,不过之前的源码确实没有了。
太忙(懒)了。
最近抽时间把平时用的医学图像分割代码整理了一下,这里以2019年的心脏数据集MS-CMRseg2019为例,提供了自己平时积累的一些源码Pytorch-medical-image-segmentation源码。
不要吐槽我的代码风格,请参考食用,喜欢的话欢迎star^ ^。
本文来自互联网用户投稿,文章观点仅代表作者本人,不代表本站立场,不承担相关法律责任。如若转载,请注明出处。 如若内容造成侵权/违法违规/事实不符,请点击【内容举报】进行投诉反馈!